StudentShare


Our website is a unique platform where students can share their papers in a matter of giving an example of the work to be done. If you find papers
matching your topic, you may use them only as an example of work. This is 100% legal. You may not submit downloaded papers as your own, that is cheating. Also you
should remember, that this work was alredy submitted once by a student who originally wrote it.
Login
Create an Account
The service is 100% legal
- Home
- Free Samples
- Premium Essays
- Editing Services
- Extra Tools
- Essay Writing Help
- About Us
✕
- Studentshare
- Subjects
- Biology
- The Promise of Protein Microarray Technology
Free
The Promise of Protein Microarray Technology - Research Proposal Example
Summary
This research proposal "The Promise of Protein Microarray Technology" discusses the generation of protein targets and ligands that will arise more possibilities of using this technique in protein research. Two major fields will be most benefitted, proteomic research and diagnostic applications…
Download full paper File format: .doc, available for editing
GRAB THE BEST PAPER94.5% of users find it useful
- Subject: Biology
- Type: Research Proposal
- Level: Undergraduate
- Pages: 4 (1000 words)
- Downloads: 0
- Author: klockocyrus
Extract of sample "The Promise of Protein Microarray Technology"
The Promise of Protein Microarray Technology Introduction. The requirement of a technology in the fast changing scientific world is in a highly parallel quantization of specific proteins in a rapid, low-cost and low-sample volume format. This has become increasing evitable with the growing recognition of the importance of the global approach to molecular characterization of physiology, development and disease. Quantification of multiple proteins simultaneously has wide application in basic biological research, molecular classification and diagnosis of disease, identification of therapeutic markers and targets, and profiling to toxin and pharmaceuticals. Parallel detection by microtiter plates, separating and visualizing proteins by two-dimensional gel electrophoresis followed by identification by mass-spectrometry are already known and is in practice but all has its own limitation. Microarray based assays using DNA-DNA interactions are well known and the protein based microarrays are just becoming popular, since there are large number of discoveries that discusses nucleic acid-protein, protein-protein, ligand-receptor and enzyme-substrate interactions. DNA-protein interactions in microarray were studied by Bulyak et al., 1999. He studied the microarrays of double stranded oligonucleotides Affymetrix technology, CA, USA is responsible for synthesizing high-density microarrays. The single stranded oligonucleotide microarray was converted into a double stranded oligonucleotide microarray by an enzymatic extension reaction.
Body
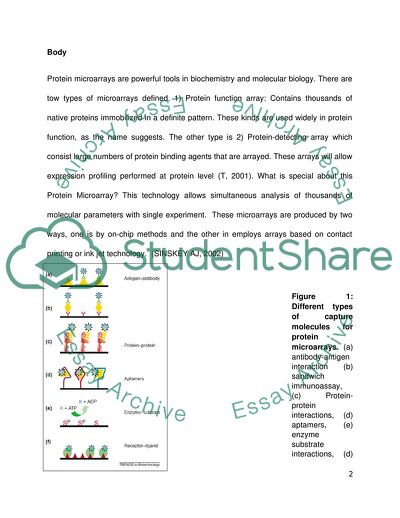
Protein microarrays are powerful tools in biochemistry and molecular biology. There are tow types of microarrays defined. 1) Protein function array: Contains thousands of native proteins immobilized in a definite pattern. These kinds are used widely in protein function, as the name suggests. The other type is 2) Protein-detecting array which consist large numbers of protein binding agents that are arrayed. These arrays will allow expression profiling performed at protein level (T, 2001). What is special about this Protein Microarray? This technology allows simultaneous analysis of thousands of molecular parameters with single experiment. These microarrays are produced by two ways, one is by on-chip methods and the other in employs arrays based on contact printing or ink jet technology. (SINSKEY AJ, 2002)
Figure 1: Different types of capture molecules for protein microarrays. (a) antibody-antigen interaction (b) sandwich immunoassay, (c) Protein-protein interactions, (d) aptamers, (e) enzyme substrate interactions, (d) receptor –ligand interactions. (Markus F. Templin, 2002)
There is tremendous importance of protein microarrays for the researchers since it paves a way to address questions at various levels by studying the interactions between protein-protein, antigen-antibody, enzyme-substrate, protein-DNA and ligand receptor. A recent implication of this technology was published. The researchers demonstrated the extraordinary powers of array based method for proteomic purposes. Different recombinant proteins (5800) from S.cerevisae were purified and the researchers generated complex proteome chips that contained gene-products from more than 90% of the genes form the yeast. These microarrays could be used to study protein-protein interactions on a genome-wide scale. The researchers used calmodulim as a model protein to probe the arrays, they could confirm many known interactions and also showed that the identification of proteins that are able to bind to low molecular weights compounds is possible. The microarray technology is also used be used diagnostic purposes in which several parameters of alone sample have to be analyzed in parallel. Figure 1 displays the use of microarray technology to screen for antigen-antibody reactions. Sandwich assays are also miniaturized and parallelized and performed in a microarray format. This research particularly opens the possibility to examine an entire proteome directly for protein-drug interactions. Accurate quantification with protein microarrays can be very well achieved, if appropriate positive and negative control spots and/or internal calibration spots are used, which will lead to robust and reliable diagnostic assays. (Markus F. Templin, 2002)
Haab BB et al., 2001 utilized protein microarrays for highly parallel detection and quantification of specific proteins and antibodies in complex solutions. This array provided specific binding sites for proteins that one wishes to measure in complex samples. The specificity, sensitivity and accuracy of the assay was analyzed in performing 115 antibody/antigen pairs. Fifty percent of the arrayed antigens and 20% of the arrayed antibodies provided specific and accurate measurements of their respective ligands at micrograms concentration levels. The authors also concluded that the protein microarrays provide a practical means to characterize patters of variation of thousands of different proteins in clinical and research applications. (Haab BB, 2001)
Limitations:
Today we have reached from DNA microarrays to protein microarrays, but technical hurdles still remains. One is the source of capture molecules and dealing with the proteins. To start an experiment using protein microarrays a starter will require a rapid high-quality isolation techniques. The specific assay questions have to be designed and the scientists need reliable ways to measure formation and well as dissociation, of protein purifications. Proteins have varying physical/chemical properties; these are subjectable to changes when the protein undergoes post-translational modification. Moreover, the proteins on the microarray should retain their functional state. Post-translation and cellular compartmentalization of proteins also poses challenges. Moreover, the proteins have variable half-lives that make the constructing protein microarrays more challenging. In the recent years, protein microarray technology is already a useful tool to study different kinds of protein interactions. (Sinskey AJ, 2002)
Future Prospects:
Further research and developments with high-throughput generation of protein targets and ligands will arise more possibilities of using this technique in protein research. Two major fields will be most benefitted, proteomic research and diagnostic applications. In medical research, it will increase the immune diagnostics significantly analyzing all the parameters of interest. Most importantly, it will require less sample volume thus minimal amounts of sample will be available like the analysis of multiple tumor markers for minima amount of biopsy material. It also opens up a new possibility of monitoring the patients during their treatments. Microarrays along with DAN chips will also increase the basic research in the area of protein-protein interactions and thus also opening possibility of protein profiling form the limited number of proteins up to high density based proteomic approaches. Enzyme specificity can also be measured by the use of protein and peptide microarrays and also for the measurements of enzyme activities on various substrates in parallel manner. (Markus F. Templin, 2002)
As years are passing by, we have witnessed the whole field of protein array technology showing a dynamic development driven by the accelerating genomic information. Automated protein expression and purification systems for the capture molecules and the analysis of whole proteomes will lead to major developments within the technology of microarray. Things with protein microarrays have just begun!
Bibliography
Haab BB, D. M. (2001). Protein MCIroarrays for highly parallel detection and quatitation of specific proteins and antibodies in complex solutions. Genome Biology , 1-13.
Markus F. Templin, D. S. (2002). Protein microarray technology. TRENDS in Biotechnology Vol.20 No.4 April 2002 , 130-166.
SINSKEY AJ, f. s. (2002). The promise of Protein Microarray. PharmaGenomics , 20-23.
T, K. (2001). Protein Microarrays: prospects and problems. Chemistry and Biology , 105-115.
Read
More
CHECK THESE SAMPLES OF The Promise of Protein Microarray Technology
Have Proteomic Approaches Advanced the Field of Cancer Biomarkers
Resources are being put together at speed in all-around technology, software, and physics, biotechnology, and chemistry in a bid to lower the mortality rate.... The advantage of proteomic technology is borne in the fact that the protein itself is the highest point of expression of these protein activities.... Some of the known various forms of cancer biomarkers include protein subgroups like enzymes, receptors, and glycoproteins along with hormones....
8 Pages
(2000 words)
Literature review
Using Tiling Arrays to Diagnose Drug Resistance in Clinical Isolates of Gonorrhea
iling array is a derivation of microarray technology developed by Kapranov and colleagues (2002) and Shoemaker and colleagues (2001) that facilitates identification of previously unidentified transcripts through genome wide annotations.... The protocol is based on similar technology already used for development of protocols for drug resistance in other organisms....
17 Pages
(4250 words)
Essay
The Meta-analysis of Breast Cancer Microarray Studies
This review examines an article regarding a meta-analysis of breast cancer microarray studies in conjunction with conserved cis-elements suggests patterns for coordinate regulation.... The writer would evaluate the effectiveness of the chosen approach and analyze the results.... ...
12 Pages
(3000 words)
Literature review
Proteomic Approaches and Reviews on Cancer Biomarker Researches
The author outlines round technology, software, and physics, biotechnology, and chemistry in a bid to lower the mortality rate.... The advantage of proteomic technology is borne in the fact that the protein itself is the highest point of expression of these protein activities.... Some of the known various forms of cancer biomarkers include protein subgroups like enzymes, receptors, and glycoproteins along with hormones.... Proteomic knowledge refers to knowledge that pertains to the protein content of the cell proteomics allow for splitting of the various mRNAs and expressing how the genes and soft genes of regulation are in contrast with genomes, it is more dynamic than the genome and consists of other forms of gene expression involving....
8 Pages
(2000 words)
Literature review
Analysis of Proteomics Problems
Formalin-fixed, paraffin wax-embedded tissue for multiple tumor tissue microarray constructions is normally used.... The author of the "Analysis of Proteomics Problems" paper identifies a methodology for a phosphoprotein of interest, describes protein interactions to detect Signaling Pathways, and appraisal of high-throughput techniques to identify and classify tumor types.... The protein separated by electrophoresis is immobilized on the hydrophobic membrane and is brought near the antibody and the phosphoprotein acts as its target antigen which forms an antigen-antibody complex by any type of antigen-antibody interactions....
10 Pages
(2500 words)
Assignment
Omic Technologies in Cancer Diseases
In the past, genes were examined independently but microarray technology has brought significant changes in recent years.... The author outlines that this technology has been pragmatic to different segments of CHM investigation starting from regularization and eminence regulation of herbal formularies, characterization of target mediated, and downstream effects.... This technology is mainly aimed at the universal detection of metabolites mRNA genes, and proteins (proteomics) in a specific biological sample in a non-biased and....
6 Pages
(1500 words)
Essay
Lymphoma Diagnostics Techniques
D DNA microarray studies have highlighted molecular pathways that are important in cancer subclasses.... The paper "Lymphoma Diagnostics Techniques" highlights that generally speaking, equipping health centers with the necessary equipment that can be used to improve human health status will help to overcome the problem of inaccessibility in remote areas....
12 Pages
(3000 words)
Essay
Regulation of Flavonoid Biosynthesis During Tomato Fruit Ripening
imple diploid genetics, small genome size, short generation time, routine transformation technology, and availability of genetic and genomic resources, including mapping populations, mapped DNA markers.... From the paper "Regulation of Flavonoid Biosynthesis During Tomato Fruit Ripening", flavonoids are polyphenolic compounds that are ubiquitous in nature and are categorized, according to chemical structure, into flavonols, flavones, flavanones, isoflavones, catechins, anthocyanidins, and chalcones....
7 Pages
(1750 words)
Research Paper
sponsored ads
Save Your Time for More Important Things
Let us write or edit the research proposal on your topic
"The Promise of Protein Microarray Technology"
with a personal 20% discount.
GRAB THE BEST PAPER

✕
- TERMS & CONDITIONS
- PRIVACY POLICY
- COOKIES POLICY